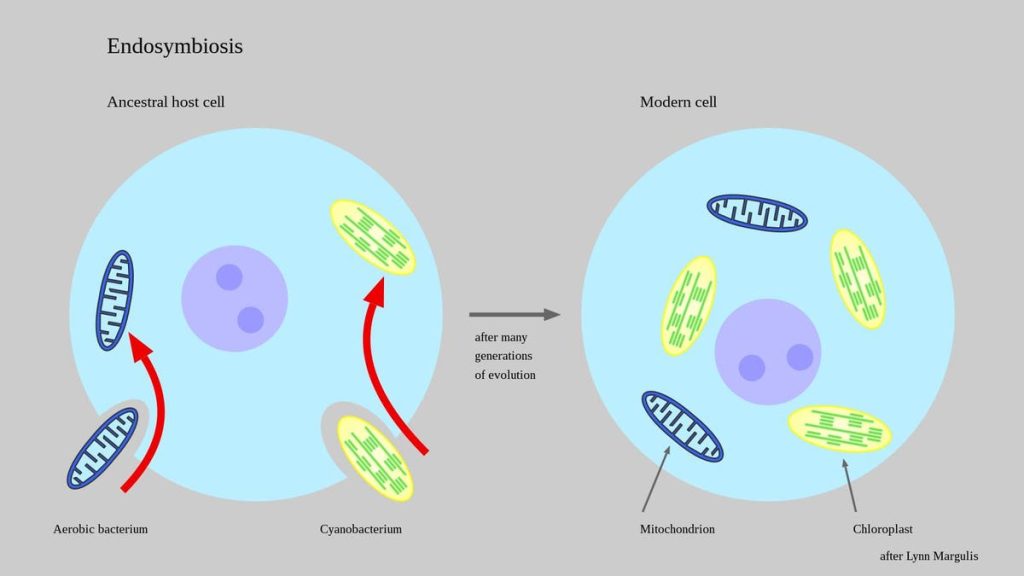
Marine bacteria found in hot springs may be the closest living relatives of the ancient bacteria that gave rise to modern mitochondria more than a billion years ago
© Copyright by GrrlScientist | hosted by Forbes | LinkTr.ee
Life on Earth started out as independent microscopic cells around 3.7 billion years ago. And so they persisted for aeons until one fateful day around 1.6-1.8 billion years ago when two ancient microbes met and created a novel partnership never before seen on Earth. One of those ancient microbes was an α-proteobacteria, a member of an extremely abundant and diverse group of bacteria that carry out all sorts of useful biochemical reactions. (The Rickettsia, for example, are members of this taxonomic class of bacteria. They should be familiar to you because they include highly pathogenic microbes such as those that cause Rocky Mountain Spotted Fever.)
But not all α-proteobacteria are deadly. Some of them are downright cooperative. One particularly helpful microbe was the ancient ancestor of modern mitochondria, those critically important metabolic “powerhouses” residing inside almost all cells containing a nucleus (known as eukaryotic cells) that make up all multicellular living beings.
Because this microbial merger occurred so very long ago, our knowledge of the details of the bacterial origin and identity of mitochondria’s ancient ancestor remains fuzzy. Additionally, microbes aren’t dinosaurs; they don’t leave fossils in stones for scientists to study and ponder, so we must infer how and when this fortuitous event occurred by using a variety of biochemical, genetic and cellular methodologies.
This microbial merger was critically important. Not only did it trigger the evolutionary burst of all multicellular life forms that have ever lived on Earth, but it influenced the most intimate aspects of our lives, including the biochemical pathways used to break down molecules to generate energy, which then fuel biosynthetic processes necessary to carry out mechanical work. For these reasons, the identity of this last eukaryotic common ancestor (LECA) that gave rise to modern eukaryotic cells containing mitochondria attracts a lot of curiosity.
To begin this study, it was necessary to identify precisely which of the myriads of α-proteobacteria could potentially be the closest relatives to the ancestors to mitochondria. This was the question confronting biochemist Otto Geiger, a professor of ecological genomics at the National Autonomous University of Mexico, and an editor for the scientific journal, BMC Microbiology, and for the Handbook of Hydrocarbon and Lipid Microbiology.
In this new study, Professor Geiger assembled a team of collaborators and together, they used a variety of biochemical, genetic and cellular approaches to narrow down the field of candidates to a small group of microbes that might potentially be the closest living relatives of the ancestral bacteria that gave rise to mitochondria.
To do this, Professor Geiger and collaborators identified a dozen metabolic traits that they thought were most likely to be present in the ancestral proto-mitochondria. They then surveyed all known genomes of α-proteobacteria — literally thousands of them — and compared their traits to those that most closely match those they had chosen their chosen.
But there is a wrinkle in this strategy: bacteria are tricky because they engage in lateral gene transfer by shuffling and redistributing genes from one species and genome to another, somewhat like shuffling and dealing out a deck of cards. Over evolutionary time, this shuffling creates new combinations and collections of genes and biochemical capabilities across different lineages such that no single α-proteobacterial lineage possesses the same collection of genes (and traits) present in the ancestral proto-mitochondria 1.5 billion years ago.
A defining step in this survey was when Professor Geiger and collaborators focused on the ability of candidate ancestral α-proteobacteria to manufacture two types of fatty compounds, known as lipids, that are essential to proper functioning of all mitochondria. These lipids are special because they are only found in a few types of modern bacteria — and this narrowed the search considerably.
One of these lipids, called ceramide, is special because it is only manufactured by mitochondria. The other key lipid, cardiolipin, is involved in respiration and energy production in mitochondria.
The presence of the genes necessary to manufacture both lipids was important because together they additionally play a critical role in helping to signal when mitochondria are damaged and need to be removed. This essential function is conserved in all eukaryotic cells, Professor Geiger said, suggesting that the candidate proto-mitochondria also possessed genes to produce both lipids.
But which modern α-proteobacteria possess the genes necessary to manufacture both lipids?
Professor Geiger and collaborators discovered these traits in free-living marine α-proteobacteria that are common in hot springs around the world. Interestingly, this group of bacteria had never before been considered as the evolutionary progenitors of modern mitochondria.
Professor Geiger and collaborators suggest these marine α-proteobacteria, belonging to the Iodidimonadales genus, are good candidates to represent modern relatives of the ancient ancestral mitochondrial bacteria because they possess traits that he and his collaborators most associated with the proto-mitochondria. Further, another factor that ties all the Iodidimonadales together, the team point out, is the high oxygen gradient in the places where they live and need to survive.
“These bacteria really depend on oxygen,” Professor Gieger concluded, in the same way that mitochondria do to produce energy.
Sources:
Otto Geiger, Alejandro Sanchez-Flores, Jonathan Padilla-Gomez, and Mauro Degli Esposti (2023). Multiple approaches of cellular metabolism define the bacterial ancestry of mitochondria, Science Advances 9(32) | doi:10.1126/sciadv.adh0066
Parth K. Raval, William F. Martin, and Sven B. Gould (2023). Mitochondrial evolution: Gene shuffling, endosymbiosis, and signaling, Science Advances 9(32) | doi:10.1126/sciadv.adj4493
SHA-256: 9ab94921e06b203a216cb219d873f92ea4083642075e2e0be632939cd42949aa
Socials: Bluesky | CounterSocial | LinkedIn | Mastodon | MeWe | Post.News | Spoutible | SubStack | Tribel | Tumblr | Twitter
Read the full article here